STRUCTURES Blog > Posts > Structure Formation on Command in Biological Cells
Structure Formation on Command in Biological Cells
This blog post is based on a transcript of the talk “Strukturbildung auf Kommando in biologischen Zellen” given by Prof Ulrich Schwarz at the public lecture series event “Akademische Mittagspause 2023: Strukturen in der Welt”, and published with kind permission. Parts of the transcript have been edited with the help of the artificial intelligence (AI) tools DeepL and ChatGPT before the text was finally copy-edited by the STRUCTURES Blog Editorial Board together with Ulrich Schwarz.
In this blog post, we will explore the fascinating world of structure formation in biological cells, which lies at the intersection of physics and biology. In biological systems, molecules like lipids, proteins and nucleic acids interact to create intricate and functional structures. Let’s delve into the significance of cells, their composition, and the remarkable processes of structure formation taking place within them.
The Story Behind Cells
Cells, the fundamental building blocks of life, are remarkably diverse and complex entities. They make up all living organisms, ranging from bacteria to complex multicellular organisms like plants and animals.
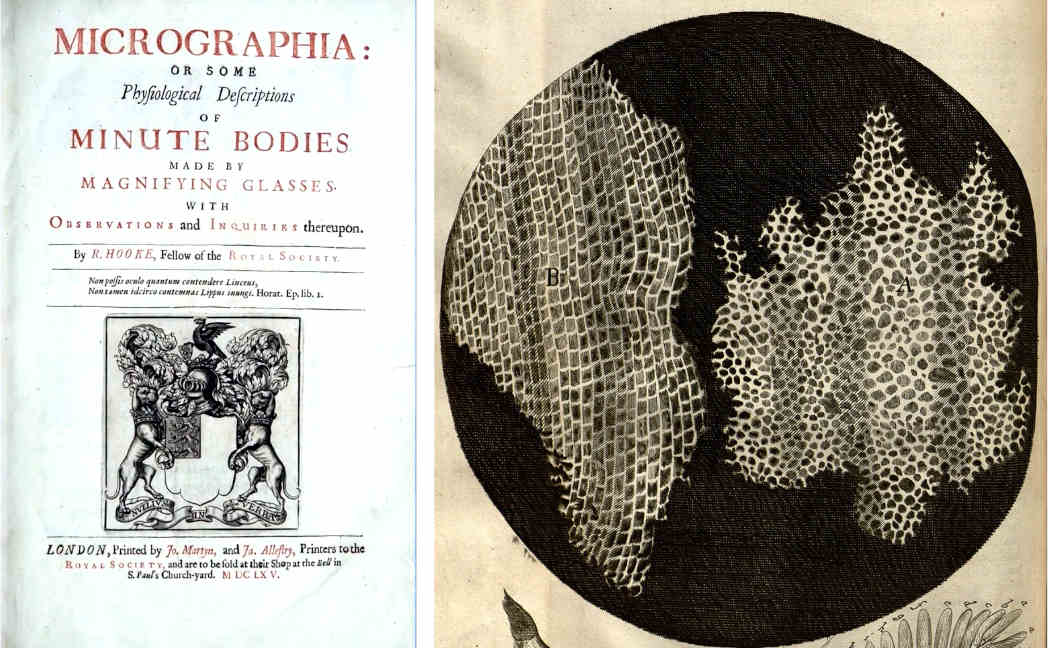
The term “cell” originated from the observations of Robert Hooke, a 17th-century scientist who used microscopes to explore the world of small objects. Through his microscope, Hooke observed compartments within cork tissue that resembled the small rooms where monks resided, leading him to coin the term cell. This discovery paved the way for the realization that all forms of life are composed of cells, which arise through the division of pre-existing cells.
The Abundance, Size and Interior of Cells
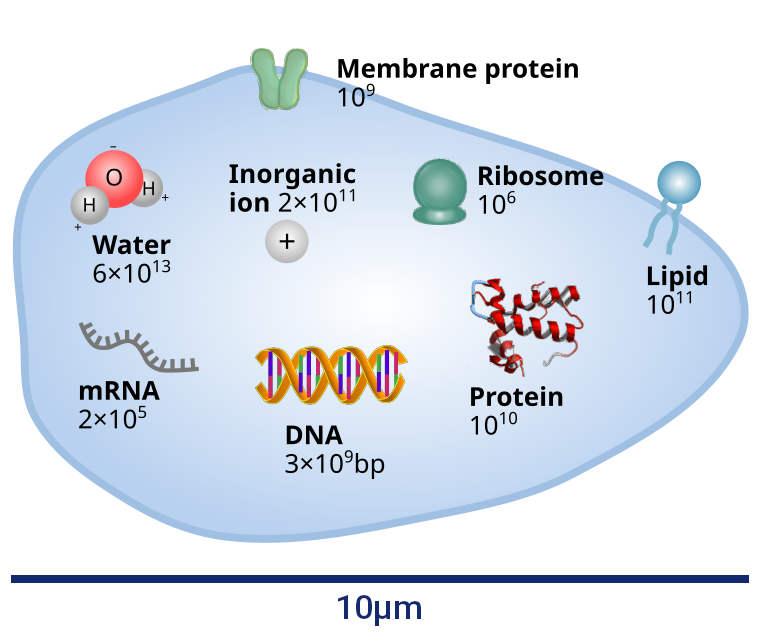
The human body is composed of an astounding number of cells, estimated to be around $3 \times 10^{13}$. This staggering quantity exceeds our imagination, highlighting the complex nature of our cellular composition. While each cell is minute, their collective presence shapes our existence and understanding the quantity of cells provides valuable insight into our cellular nature. By considering the volume of an average human body and dividing it by the number of cells, we immediately obtain a rough estimate of the typical size of a cell, arriving at a length scale of approximately $10 \mu m$. This value indeed aligns with what we observe under a microscope, where cells exhibit a typical size on the order of $10 \mu m$ – a size that allows for easy observation under an optical microscope.
To understand structure formation in cells, let’s take a step inside and examine their interior content. The primary component within cells is water, accounting for 70 percent of our body and consisting of approximately $6 \times 10^{13}$ water molecules per cell. Water provides the essential aqueous environment for the proper functioning of biomolecules within cells, acting as a solvent for their activities. Apart from water, cells contain a “dry mass” dominated by proteins. Proteins are essentially molecular workhorses that carry out crucial tasks such as enzymatic reactions, nutrient transport, hormonal regulation, and antibody production. While lipids, RNA, and DNA also contribute to the intricate machinery of the cell, our focus will primarily be on proteins, which amount to approximately $10^{10}$ per human cell. Considering the volume of a cell, this translates to a typical protein concentration on the order of millimolar, corresponding to an average intermolecular distance of about 10 nanometres. Thus, cells exhibit a relatively high density of proteins.
The “Dance of Life”
Within the crowded cellular environments, proteins and other molecules face challenges related to diffusion and spatial organization. The high density of cellular components slows down the molecules, hindering their movement compared to dilute solutions. However, this high density also promotes frequent encounters and interactions between molecules, enabling efficient signalling, enzyme activity, and other vital cellular processes. Thus, the density of proteins in a cell provides a careful balance: it is just high enough to (still) allow movement, while at the same time maintaining a large number of interactions that is necessary in order to be able to exchange and transmit information.
For Advanced Readers: The Physics of Protein Motion
Molecules are constantly in motion. The physics of this motion works just as for any other (non-bio)molecules. The movement of proteins relies on two basic laws:
- The equipartition theorem states that kinetic energy is determined by temperature:
$ \begin{align} \frac 12 m v^2 = \frac{k_B T}2 \Rightarrow v = 10 \frac ms \end{align} $
By applying typical values for mass and temperature, we can calculate velocities of about 10 metres per second for proteins. This is rather high, resulting in permanent collisions of the molecules. These interactions can include binding to specific targets, undergoing conformational changes, or participating in larger-scale assembly processes.
- The Stokes-Einstein relation: As the molecule is moving on a “zig-zag” course through the cell due to its many collisions, its motion represents a diffusion process that is characterized by a diffusion constant “$D$”, which again is determined primarily by the (body or room) temperature:
$ \begin{align} D = \frac{k_B T}{6 \pi \eta R} \Rightarrow D = \frac {(10 \mu m)^2}s \end{align} $
This means that a biomolecule needs one second to cross a cell of a size of $10 \mu m$, on its way interacting with all the other molecules.
It is important to understand the necessity of high molecular densities within cells for comprehending their biochemistry. The exchange of information among molecules relies on numerous interactions, which can only occur effectively in densely packed environments. This concept sheds light on why cells typically have a limited size of around $10 \mu m$. If cells were significantly larger, the distances would become too vast. Given that the diffusion parameters are fixed by physical laws, the time required to transmit information would increase significantly, rendering the cell unable to make efficient decisions. To enable information exchange on larger scales, specialized mechanisms like nerve conduits come into play, as they provide unique pathways for communication.
From Physics to Biology: The Formation of Structures within Cells
Within the microscopic world of cells, the densities of molecules play a vital role in facilitating efficient biochemical processes. However, high densities can also pose challenges. In principle, they can give rise to the formation of harmful structures. For example, it could happen that the concentration of certain components reaches excessive levels, compelling their molecules to solidify and leading to the development of gallstones or bladder stones. To prevent such detrimental outcomes, cells must carefully regulate the process of structure formation, ensuring it does not spiral out of control and avoids any pathological consequences.
One of the remarkable aspects of cellular biology therefore is the ability of cells to self-organize. The formation of structures is tightly regulated and controlled by the cell, ensuring that it occurs only “on demand”, i.e. when it is necessary, while avoiding any unwanted outcomes. This process involves the precise arrangement of molecules and organelles. Signalling pathways, gene expression, and feedback loops work in harmony to govern the timing, location, and extent of structure formation. Any disruption to these regulatory mechanisms can lead to cellular dysfunction and disease, underscoring the importance of precise control in maintaining cellular integrity.
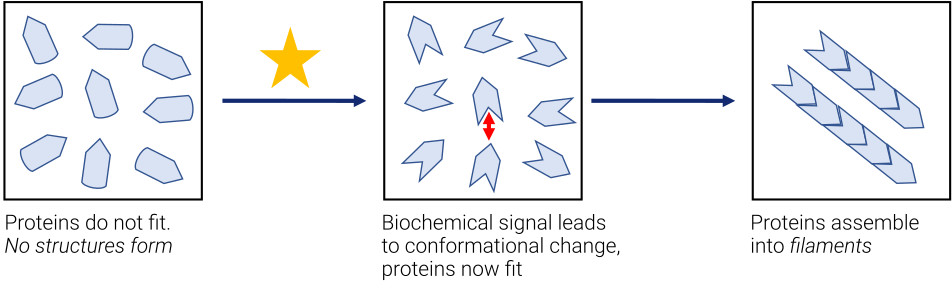
To better understand the principles at play, imagine a box containing proteins that have a specific shape, as in Fig. 4. In the left panel, the proteins resemble puzzle pieces that fail to fit together due to their concave, pointed and rounded shapes. However, when a biochemical signal arrives and adjusts the properties of proteins (see the middle and right panels), the pieces fit together and can now align to form a larger object. We refer to these two states as “low affinity” and “high affinity”, respectively. Initially, the proteins have no affinity for each other, but with the appropriate conformation, structure formation can take place. In this simple case, it would result in multiple proteins coming together to form filaments. The key lies in the biochemical signal, which ensures a high affinity, leading to subsequent crystal formations. This process occurs continuously within our cells.
Phase Separations in Physics and Biology
How do structures form in physics? A valuable approach is to examine the phase behaviour of simple systems, such as for instance a Lennard-Jones fluid. A Lennard-Jones fluid refers to a theoretical model used to describe the behavior of a collection of fluid particles. It is named after physicist John Lennard-Jones, who proposed the model in 1924, and describes the intermolecular interactions between the fluid particles.
Lennard-Jones fluid
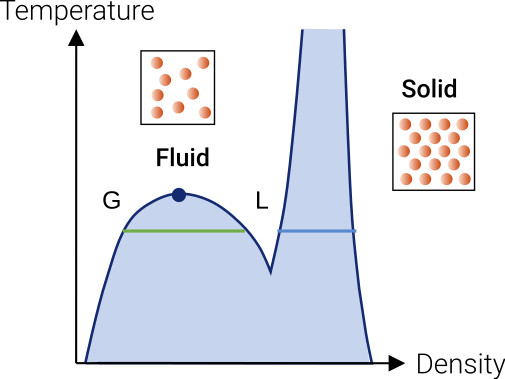
Fig. 5 illustrates a so-called phase diagram of a Lennard-Jones fluid, depicting how the system behaves under different temperature and density conditions. On the right side of the diagram, at higher densities, the system solidifies, analogous to the formation of gall or bladder stones when molecular concentrations force an ordered lattice arrangement. At lower densities, the system remains unstructured, resembling a fluid lacking long-range order but exhibiting short-range order. These distinct phases result in vastly different macroscopic forms – leading to either crystals with defined edges or rounded shapes like water or oil droplets.
Between these two phases, there exist transitions. As a high-temperature fluid cools down, it may undergo a phenomenon known as fluid-fluid phase separation. This means that the fluid splits into both a liquid and a gas phase. The fluid-solid phase transition occurs when density is increased.
After this brief excursion to physics, let’s return to the biological realm, which essentially encompasses physics with biomolecules. Here, from the distinctive characteristics of proteins, the same two phase transitions, known from physics, arise:
-
First, we have a fluid-fluid phase separation, which in the biological context is called a liquid-liquid phase separation (or in short LLPS), and initially was believed not to exist in biology until the discovery of droplet-shaped structures in cells around 14 years ago. These droplets exhibit properties characteristic of liquid droplets, including short-range order. Today, we know that proteins and nucleic acids often localize within these liquid droplets, a phenomenon facilitated by a special type of proteins that was not known until recently: intrinsically disordered proteins (IDPs), proteins with non-ordered sequences. Cliff Brangwynne and colleagues [4] first described the formation of droplet-shaped structures, demonstrating their existence in fertilized egg cells and their enrichment in essential substances for embryo development.
-
The second possibility is the fluid-solid phase transition, which has been an area of extensive study since it involves the well-known type of folded proteins. Through a process known as “self-assembly”, these proteins form so-called protein complexes. These complexes adopting specific configurations, enabling the formation of crystal-like structures with defined edges.
Structure formation on command – “remote controlling” cells
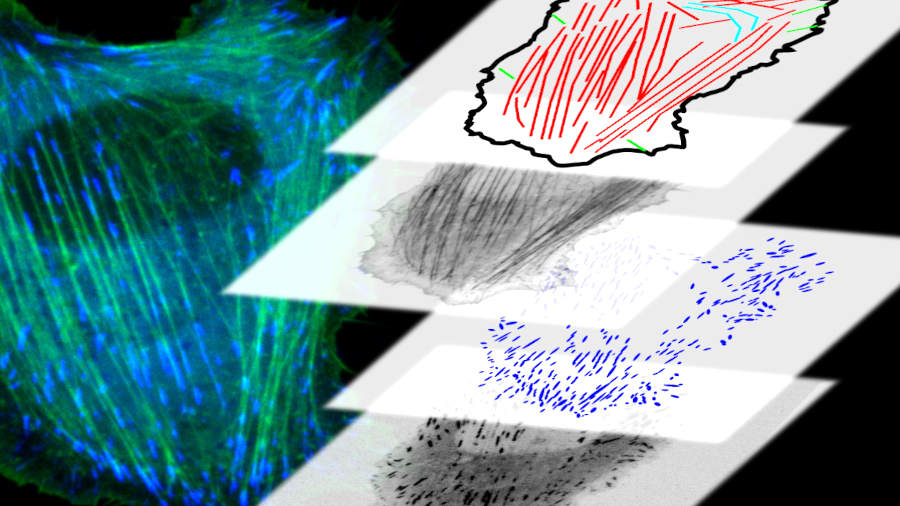
The self-assembly of protein complexes is an intriguing example for the self-organized process of structure formation within cells. While cells in a solution within a test tube exhibit relatively few structures (mainly the cell nucleus and some membrane-based structures), when placing the cells on a surface, complex and interesting structures form within minutes. The biochemical signal induced due to the contact with the surface instructs the cell to organize itself differently, necessitating the formation of numerous structures. Fig. 6 (lower row) illustrates this phenomenon, showing the cytoskeleton on the left, adhesion contacts in the middle, and the forces calculated for these structures on the right. We can observe an unordered polymer network at the top of the cell, while a well-defined structure forms at the bottom, directly interacting with the surface and aiding the cell’s adhesion. These structures have formed within 10 minutes.
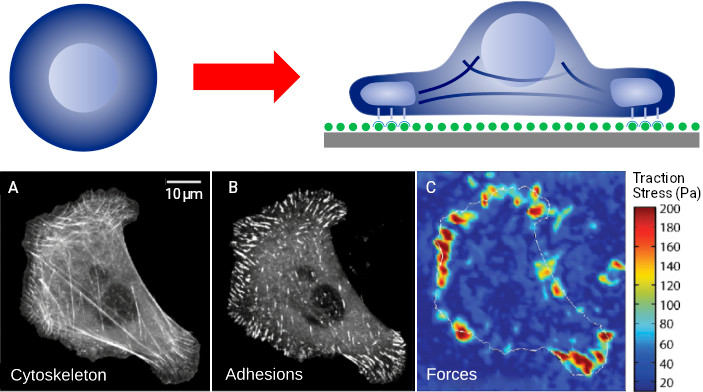
What makes this even more fascinating is that once we understand the commands that trigger structure formation in cells, we can exert control and manipulate the system. Optogenetics, a field that harnesses light-sensitive genetically engineered molecules, allows for biochemical control of structure formation from outside the cell. By shining light on the cell, we can activate these “signal molecules” and remotely guide structure formation, akin to having a “remote control” for cellular processes.
To summarize, the formation of structures within biological cells is a fascinating phenomenon that involves intricate arrangements of biomolecules (mainly proteins). Cells possess the remarkable ability to self-organize, creating dense yet mobile environments where molecules diffuse, interact, and generate structures as needed. By unravelling the mysteries of structure formation in cells, we gain deeper insights into the complexities of life itself.
Furthermore, this knowledge has wide-ranging implications for fields such as biotechnology, medicine, and synthetic biology. Unravelling the mysteries of structure formation in cells not only deepens our understanding of life’s complexities, but also opens doors to innovative applications and advancements in various scientific disciplines.
References
- Soiné JRD, Brand CA, Stricker J, Oakes PW, Gardel ML, Schwarz US (2015) Model-based Traction Force Microscopy Reveals Differential Tension in Cellular Actin Bundles. PLoS Computational Biology 11(3): e1004076. https://doi.org/10.1371/journal.pcbi.1004076
- Milo, R., & Phillips, R. (2015). Cell biology by the numbers. Garland Science.
- McGuffee, S. R. and Elcock, A. H., “Diffusion, Crowding & Protein Stability in a Dynamic Molecular Model of the Bacterial Cytoplasm”, PLoS Computational Biology, vol. 6, no. 3, p. e1000694, 2010. https://doi.org/10.1371/journal.pcbi.1000694.
- Brangwynne, C. P., “Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation”, Science, vol. 324, no. 5935, p. 1729, 2009. https://doi.org/10.1126/science.1172046.
- Schwarz, U. S., & Gardel, M. L. (2012). United we stand–integrating the actin cytoskeleton and cell–matrix adhesions in cellular mechanotransduction. Journal of cell science, 125(13), 3051-3060. https://doi.org/10.1242/jcs.093716
Tags:
Structure formation
Biophysics
Cells
Molecules
Self-Organization
Diffusion
Phase Transitions